Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer
Wilson lab paper featured in Nature Communications.
Authors
Wilson, M. D., Renault, L., Maskell, D. P., Ghoneim, M., Pye, V. E., Nans, A., Rueda, D. S., Cherepanov, P., & Costa, A.
Summary of Paper by Lori Koch
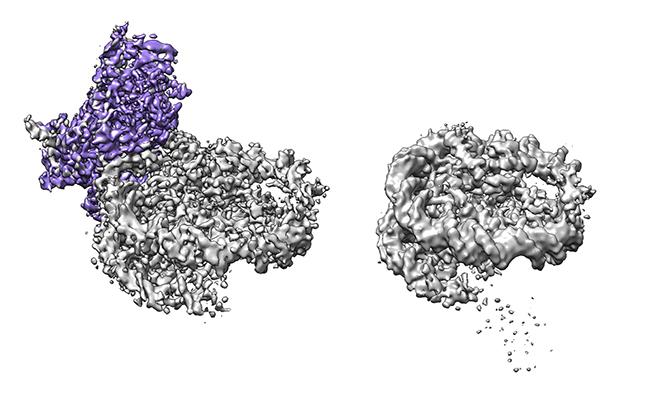
Retroviruses must integrate into the host DNA in order to reproduce. In cells, DNA is wrapped around protein complexes called nucleosomes that can shift and slide to allow protein factors to access the DNA. Previous work showed that the model retrovirus PFV integrase complex, called the intasome, interacts with the human nucleosome core particle (NCP) under active integration conditions. Low resolution structures of this complex revealed how the DNA is looped out at the integration site upon viral enzyme engagement. In this publication in Nature Communications, WCB scientist Marcus Wilson aimed to uncover what additional structural changes occur during integration. To do this, the researchers used cryo-electron microscopy (EM) to determine structures of the intasome-NCP complex and nucleosome alone with more detail. The structures revealed that the N-terminal tail of the H2A protein in the nucleosome interacts with the intasome. They also verified that this does not occur in the context of a “native” NCP assembled with DNA that does not “position” the nucleosome as tightly. Strikingly, their new cryo-EM data revealed that the host DNA slides by two base-pairs during viral integration in a manner analogous to chromatin remodellers. They then confirmed the measurements using single-molecule FRET (Forster resonance energy transfer) assays. Overall, the study presents significant advances both in the methodology of visualizing native NCPs and in its findings on the structural mechanisms of retroviral integration.

