Integrative vectors for regulated expression of SARS-CoV-2 proteins implicated in RNA metabolism
Tollervey lab paper featured in Wellcome Open Research.
Authors
Bresson, S., Robertson, N., Sani, E., Turowski, T.W., Shchepachev, V., Kompauerova, M., Spanos, C., Helwak, A., and Tollervey, D.
Summary of Paper by Lori Koch
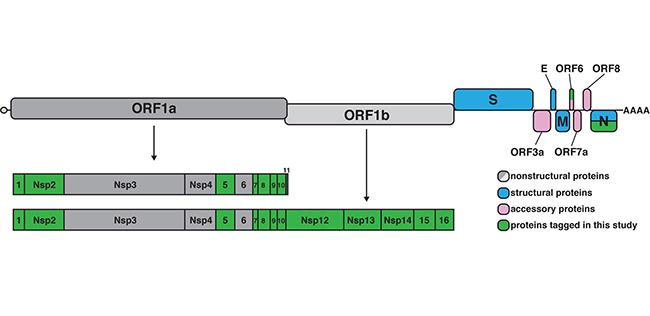
COVID-19 is caused by the SARS-CoV-2 virus which hijacks host cell RNA metabolism to replicate its long, single-stranded RNA genome. Some of the virus’s non-structural proteins interact with the host cell RNA to influence gene expression and evade the immune response. We need to better understand the interaction between viral proteins and host RNA to develop therapies. To this end, researchers in the Tollervey group led by postdoctoral scientists Stefan Bresson and Aleksandra Helwak constructed a library of viral protein expression vectors which can be used to produce SARS-CoV-2 proteins in mammalian host cells in a controlled manner. The construction of these vectors and stable cell lines was described in a report in Wellcome Open Research. The vectors place the proteins under the control of a drug-inducible promoter, which allows for varied expression according to dosage as well as time since induction. Careful control of the viral protein level is critical to ensure host-virus interactions are studied in their most relevant conditions. In the report, the scientists quantified the expression level of each of the 14 potential RNA-binding proteins using standard western blotting by comparing to an internal standard measured by mass spectrometry. Overall, the tools and methods developed in this work build the foundation for future studies. In particular, global RNA-protein interaction mapping using the Tollervey lab developed methods CRAC and TRAPP is now possible given these tools.
Related links

