In-cell architecture of an actively transcribing-translating expressome
Rappsilber lab paper featured in Science.
Authors
O’Reilly, F.J., Xue, L., Graziadei, A., Sinn, L., Lenz, S., Tegunov, D., Blötz, C., Singh, N., Hagen, W.J.H., Cramer, P., Stülke, J., Mahamid, J., and Rappsilber, J.
Summary of Paper by Lori Koch
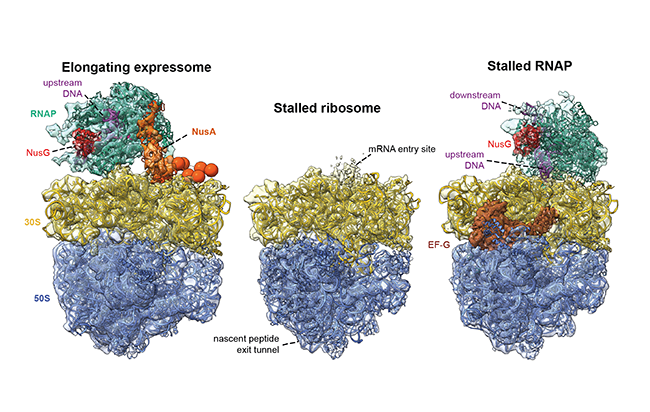
Proteins in the cell are made through the related processes of transcription and translation. During transcription, the enzyme RNA polymerase creates messenger RNA from the complementary DNA sequence and then during translation the ribosome “reads” messenger RNA and produces the appropriate protein. In bacteria cells, these processes are so closely linked that RNA polymerase and the ribosome are physically linked together and form a supercomplex known as the expressome. Previously, the expressome was reconstituted in vitro but only in a stalled state, which could not provide a mechanism for the active coupling of transcription and translation machineries observed in biochemical experiments. In their recent publication in Science, researchers led by Francis O’Reilly in the Rappsilber group (WCB/TU Berlin) determined the structure of this complex within cells at high resolution. To do this, they used a novel combination of in-cell crosslinking mass spectrometry, developed in the Rappsilber lab, and whole-cell cryo-electron tomography, developed in the Mahamid lab at EMBL Heidelberg. They were able to visualise the active expressome complex in the simple bacteria Mycoplasma pneumoniae, which is a human pathogen. The structure of the actively elongating expressome revealed that the two complexes are bridged by the protein NusA. The scientists speculate that the protein NusA may act as a sensor of ribosome position that can modulate RNA polymerase elongation. Next, they treated cells with antibiotics that inhibited either the RNA polymerase or the ribosome and found that this disrupted complex formation in different ways, suggesting that expressome formation requires active transcription and translation. In the future, the methods developed in the study may be used to capture images of other dynamic processes within the cell at close to atomic level resolution.

