Mapping the invisible chromatin transactions of prophase chromosome remodeling
Earnshaw lab paper featured in Molecular Cell.
Authors
Samejima, I., Spanos, C., Samejima, K., Rappsilber, J., Kustatscher, G., and Earnshaw, W.C.
Summary of Paper by Lori Koch
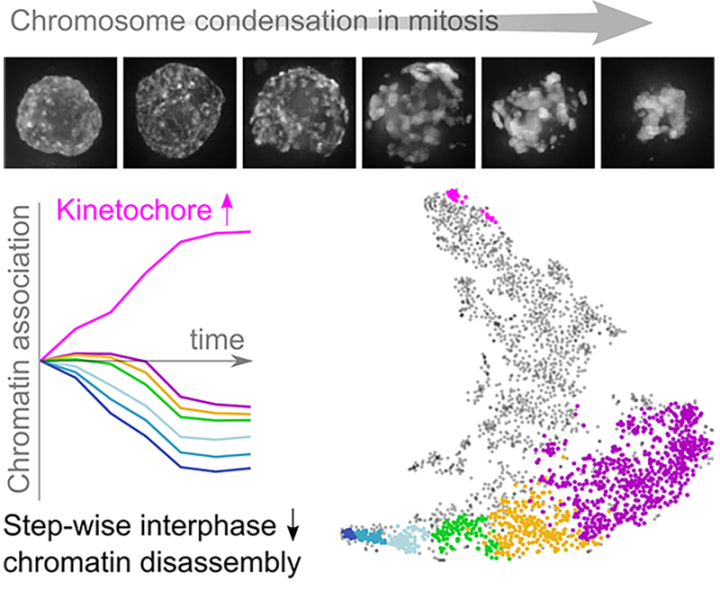
During cell division, chromosomes undergo a dramatic change in shape and composition as the interphase chromatin arrangements that allow for regulated gene expression are disassembled and the classic X shaped mitotic chromosomes form. In research published in Molecular Cell, scientists in the Earnshaw, Kustatscher and Rappsilber groups used an integrated combination of chemical genetics, proteomics and informatics to reveal, with unprecedented temporal resolution, the changes in chromatin-associated proteins from late G2 through prophase shortly after nuclear envelope breakdown. Using an engineered chicken DT40 cell line in which the major cyclin dependent kinase CDK1 can be reversibly inhibited (1), they generated populations of cells that release from G2 into prophase with >90% synchrony. Then they isolated chromatin (DNA + proteins) over a series of 5-minute intervals, up to 30 minutes after release from G2, using a method developed in the Rappsilber laboratory called ChEP (chromatin enrichment for proteomics - 2) in which proteins crosslinked to DNA are identified and quantified by SILAC mass spectrometry. They observed that ribosomal RNA processing factors were among the first proteins to leave the chromatin upon entry into prophase while RNA polymerase remained. Detachment of chromatin from the nuclear pore and from the inner nuclear envelope also occurred very early in prophase. Changes in classic chromatin proteins occurred only later. However, unexpectedly, they saw that the lamin A and lamin B proteins leave chromatin quite late. This was confirmed by live-cell imaging and immunoblotting. This study reveals the successive waves of change in chromatin proteins as the genome begins its preparations for segregation during mitosis.
Interactive app to explore the dataset
References:
(1) Samejima K, Booth DG, Ogawa H, Paulson JR, Xie L, Watson CA, Platani M, Kanemaki MT, Earnshaw WC. 2018.
(2) Kustatscher G, Wills KLH, Furlan C, Rappsilber J. 2014.
Chromatin enrichment for proteomics. Nature Protocols
Related links

